Published on: 26 January 2016 Full paper is on Plant Physiology, DOI: https://doi.org/10.1104/pp.15.01624
Abstract
Leaves are derived from heterotrophic meristem tissue that, at some point, must make the transition to autotrophy via the initiation of photosynthesis. However, the timing and spatial coordination of the molecular and cellular processes underpinning this switch are poorly characterized. Here, we report on the identification of a specific stage in rice (Oryza sativa) leaf development (P3/P4 transition) when photosynthetic competence is first established. Using a combined physiological and molecular approach, we show that elements of stomatal and vascular differentiation are coordinated with the onset of measurable light absorption for photosynthesis. Moreover, by exploring the response of the system to environmental perturbation, we show that the earliest stages of rice leaf development have significant plasticity with respect to elements of cellular differentiation of relevance for mature leaf photosynthetic performance. Finally, by performing an RNA sequencing analysis targeted at the early stages of rice leaf development, we uncover a palette of genes whose expression likely underpins the acquisition of photosynthetic capability. Our results identify the P3/P4 transition as a highly dynamic stage in rice leaf development when several processes for the initiation of photosynthetic competence are coordinated. As well as identifying gene targets for future manipulation of rice leaf structure/function, our data highlight a developmental window during which such manipulations are likely to be most effective.
Introduction
Rice (Oryza sativa) is a C3 grass whose seeds are the single most important staple food, providing one-fifth of the world’s dietary energy supply (Elert, 2014). The source of all carbon in these seeds is photosynthesis, and there is significant interest in improving this fundamental process through the alteration of several morphological, biochemical, and physiological traits in leaves (Long et al., 2006; Hibberd et al., 2008). However there are still large gaps in our knowledge of leaf development and the ontogeny of photosynthesis, in both rice and other species. For example, although we have a deep and detailed understanding of the mechanism of photosynthesis, it is still unclear at exactly which stage in development a rice leaf first gains the capacity to capture light energy for carbon fixation. Linked to this, although it is well established that leaves in many species (including rice) can undergo morphological and photosynthetic acclimation to the environment (Hikosaka and Terashima, 1995; Murchie and Horton, 1997; Oguchi et al., 2003) and it is known that the cellular processes underpinning this acclimation occur during relatively early stages of leaf development (Jurik et al., 1979; Sims and Pearcy, 1992; Murchie et al., 2005), the point when developmental plasticity is lost has not been defined precisely.
Rice leaves are initiated by a coordinated process of cell growth and division that leads to the formation of a protrusion on the shoot apical meristem (Itoh et al., 2005). The primordium (enclosed within the encircling sheaths of older leaves) then undergoes further development to form a mature leaf that consists of a flattened blade and a sheath encompassing the younger leaves, which develop sequentially from the shoot apical meristem. The young plant thus consists of a series of concentric leaves all of which originate from the meristem and are produced in a series separated by a time period, termed the plastochron (P). The definition of leaves by their sequential order of formation (L1, L2, L3…) and plastochron age (P1, P2, P3…) allows the comparison of leaves from different plants at equivalent developmental stages. P1 stage is characterized by a small protrusion forming on the flank of the shoot apical meristem. This protrusion then forms a hood-shaped structure around the shoot apical meristem (P2 stage) before completely enclosing the shoot apical meristem and taking on an elongated conical shape (P3 stage; Itoh et al., 2005). The subsequent P4 stage is characterized by a phase of rapid elongation of the leaf blade and visible greening of the tissue. Due to the degree of elongation that occurs during P4, the stage can be subdivided into stages (1–12) defined by particular blade lengths (Kusumi et al., 2010). At P5 stage, the distal tip of the leaf starts to protrude from the sheaths of older leaves, being pushed up by rapid elongation of the more proximal sheath, before growth stops as the leaf reaches maturity (P6 stage).
In addition to distinct developmental stages over time, individual grass leaves (such as rice) show a clear gradient of development along the longitudinal axis (Li et al., 2010). This has been successfully exploited in a number of investigations to allow comparisons of transcriptomes, proteomes, and metabolomes in cells undergoing different stages of differentiation. These analyses have revealed key pathways and fundamental regulators of photosynthesis in rice and also in maize (Zea mays; Li et al., 2010; Majeran et al., 2010; Pick et al., 2011; Wang et al., 2014). These patterning events at the base of mature leaves reflect a template set by the more mature differentiated tissue, distinct from the processes that must occur within a new leaf primordium, where the pattern arises de novo. Thus, studying the morphology, physiology, and gene expression dynamics of rice leaf primordia has the potential to provide novel insights that may not be evident in studies of mature leaf gradients.
With respect to the analysis of the very earliest stages of grass leaf development, previous work has successfully used laser microdissected portions of maize shoot apical meristem domains to identify transcripts associated with leaf initiation (Ohtsu et al., 2007). This work was restricted to the extremely early processes of leaf determination and initiation (up to P1) and did not encompass subsequent stages of leaf development. This was achieved by Wang et al. (2013b), who recently provided a detailed transcriptomic analysis of early leaf development in maize using dissected primordia. These data provided the first analysis of a monocot leaf at this developmental resolution. To date, a similar analysis has not been reported for rice.
Most of the work on early leaf development (in both monocots and dicots) has focused on changes in the transcriptome, yet relating these data to the physiological function of the leaf is often problematic. Gene expression at the transcript level does not necessarily imply biochemical or physiological function (Amiour et al., 2012; Fernie and Stitt, 2012; Lan et al., 2012). In the case of photosynthesis, chlorophyll fluorescence has developed as a routine technique for the measurement of photosynthetic function (Krause and Weis, 1991; Maxwell and Johnson, 2000; Meng et al., 2001; Baker, 2008); however, most equipment is designed for measurements in leaves that can be clamped within a chamber, greatly restricting the minimum size of the material that can be analyzed. Microfluorescence techniques have been developed that provide the appropriate resolution for the analysis of small leaf primordia; however, these have generally been applied to the analysis of plant-microbe interactions (for review, see Berger et al., 2007) and plant stress response (for review, see Baker and Rosenqvist, 2004) rather than used in the context of leaf development.
A significant body of work has shown that many plants have the ability to acclimate their leaves to the light environment, leading to the formation of sun or shade leaves (Kubinova, 1991; Murchie and Horton, 1997; Oguchi et al., 2003; Kim et al., 2005; Terashima et al., 2006). The ambient environment is sensed by mature leaves, leading to the formation of an as yet uncharacterized signal that influences the morphogenesis of developing leaves at a distance from the mature leaves. It is clear that this systemic signaling system exists in rice and that responding leaves are young (Murchie et al., 2005), but the precise developmental stage during which this developmental plasticity exists is not known, nor is whether this developmental window of sensitivity to acclimation correlates with the expression of particular sets of genes associated with specific elements of leaf differentiation.
Here, we report on a combined physiological and transcriptomic analysis of the early stages of rice leaf development. These data provide a description of the gene expression changes occurring during the earliest phase of acquisition of photosynthetic competence. In addition, by exploiting a series of transfer experiments between high- and low-irradiance regimes at distinct leaf stages, we identify the P3/P4 transition as a key stage in rice leaf development where a coordinated differentiation of photosynthetic tissue, vasculature, and stomata occurs.
Materials and Methods
Plant Material and Growth Conditions
Rice (Oryza sativa var indica; IR64) plants were grown in hydroponics as described (Makino et al., 1997). Plants were grown in a growth chamber (Conviron; www.conviron.com) at high light (700 µmol m⁻² s⁻¹) or low light (250 µmol m⁻² s⁻¹) with a 12-h/12-h light/dark cycle, 50% humidity, ambient CO₂, and a temperature of 28°C.
Histology
For leaf thickness measurements, 1-cm sections were cut from mature fifth leaf blades, fixed in 4:1 ethanol:acetic acid for 24 h, and embedded in Technovit 7100 (TAAB; www.taab.co.uk). Samples were sectioned at 2 µm thickness using a Leica RM2145 microtome (www.leica-microsystems.com), stained with Toluidine Blue (Sigma-Aldrich; www.sigmaaldrich.com), and imaged using an Olympus DP71 microscope (www.olympus-lifescience.com). Line drawings of vascular development were rendered manually in Adobe Photoshop CS5. Thickness was measured at the bulliform cells using Adobe Photoshop version 12.0. For transmission electron microscopy, primordia at different developmental stages were dissected into 3% (v/v) glutaraldehyde (Sigma-Aldrich) in 0.1 m phosphate buffer. Further fixation and processing were as described (Wallace et al., 2015). For confocal microscopy, P4 stage leaf primordia were dissected, mounted in water, and imaged using an inverted LSM510 Meta confocal microscope (Zeiss; www.zeiss.co.uk). Excitation was with a 488-nm argon laser. Emitted light was detected at 650 to 710 nm. Noise in the image background only was removed using Adobe Photoshop.
Scanning Electron Microscopy
Rice leaf primordia were fixed for 6 h on a 2-rpm orbital shaker in glass vials containing 4% (w/v) paraformaldehyde (Thermo Fisher Scientific), 2.5% (w/v) glutaraldehyde (Agar Scientific), 0.5% (v/v) Tween 20 (Sigma-Aldrich), and 0.2 m phosphate-buffered saline, pH 7.4 (Sigma-Aldrich), all prepared using ultrahigh purified water. Fixative was replaced with two changes of phosphate-buffered saline for 30 min each. Samples were dehydrated in acetone (Fisher Scientific) series of 5%, 10%, 25%, 40%, 55%, 70%, 85%, 95%, and two times 100% (v/v) for 1 h each. Samples were dried with liquid CO₂ using a Polaron critical point dryer (Agar Scientific). Samples were then mounted onto aluminum stubs using black carbon stickers (Agar Scientific) and sputter coated with gold (Edwards S150B gold sputter coater) in an argon gas atmosphere. Images were obtained using a scanning electron microscope (Philips XL-20) and processed with embedded XL-20 software.
Light Response Curves
Light response curves were recorded on mature fifth leaves in which blade elongation was no longer occurring. Gas exchange was recorded using an LI-6400 portable photosynthesis system (LICOR Biosciences; www.licor.com) at irradiances of 50, 100, 150, 200, 250, 300, 400, 500, 600, 800, 1,000, 1,250, 1,500, and 2,000 µmol m⁻² s⁻¹ using a constant air flow rate of 200 µmol m⁻² s⁻¹, a sample CO₂ concentration of 400 µmol m⁻² s⁻¹, a block temperature of 28°C, and a relative humidity of 50%. Plants were allowed to acclimate to each subsequent irradiance level for 3 min. The average of up to 54 photosynthetic assimilation rate measurements recorded at 5-s intervals was taken at each irradiance level.
Chlorophyll Fluorescence Microscopy
P3 and P4 stage leaf primordia were dissected, mounted on cooled set agarose in a humid environment, and imaged using a modified Olympus BX50WI microscope (Rolfe and Scholes, 2002). After exposing leaf primordia to an initial dark period of 5 min, the minimal level of fluorescence was recorded (F ₒ) and a saturating pulse was applied (3,000 μmol m⁻² s⁻¹) to measure the initial maximum fluorescence (F ₘ). Subsequently, an actinic light was switched on, and F ₛ/F ₘ′ was recorded every 30 s over a period of 20 min as primordia went through induction. Optimal induction irradiances to avoid photodamage of 50 μmol m⁻² s⁻¹ for P3 and P4 stage leaves and 200 µmol m⁻² s⁻¹ for P5 stage and mature leaves were determined through pilot experiments. For light response curves, primordia were exposed to irradiances of 30, 50, 100, 150, 200, 300, 400, and 600 μmol m⁻² s⁻¹ after undergoing induction. Plants were allowed to acclimate to each subsequent irradiance level for 5 min, after which four F ₛ/F ₘ′ measurements were taken at 30-s intervals. For induction and light response curves, data were extracted from several circular ROIs, each with a diameter of 40 µm. ROIs were evenly spaced along the entire length of the primordium (P3 and P4 stage leaves; up to seven ROIs) or positioned at the base, midpoint, and tip of P5 or mature leaf blades (three ROIs per leaf blade), with positioning over the midvein or major veins avoided (Supplemental Fig. S1C). Where no signal was recorded in multiple ROIs throughout the course of the induction or light response experiment, plotted traces may overlap. Absorbance was imaged using the red/far-red light method (Rolfe and Scholes, 2010).
Chlorophyll Content Analysis
Chlorophyll was extracted and chlorophyll content was quantified using the method described by Lichtenthaler and Wellburn (1983). Briefly, two leaf discs (each 4 mm in diameter) were collected from the middle of the P5 or mature leaf blade (n = 5 per stage) and ground up in 10 mL of cold 80% (v/v) acetone using a pestle and mortar. Chlorophyll a and b concentrations were measured using a spectrophotometer (Perkin Elmer), with the following calculations used: chlorophyll a (µg mL⁻¹ ) = 12.21 (A₆₆₃) − 2.81 (A₆₄₆); chlorophyll b (µg mL⁻¹) = 20.13 (A₆₄₆) − 5.03 (A₆₆₃).
Analysis of Stomatal and Vein Patterning
For each rice leaf, a 1-cm segment at the midpoint was cut and fixed in ethanol:acetic acid (7:1, v/v) for 2 to 3 d at room temperature, then bleached (25% [v/v] economic bleach; Ottimo Supplies) for 1 d. Samples were cleared using two to three drops of chloral hydrate solution (10 g of chloral hydrate in 2.5 mL of water and 1 g of glycerol) for 1 h at room temperature. One to two drops of modified Hoyer’s solution (10 g of chloral hydrate from Riedel-de Haen, 1 g of glycerol from Sigma-Aldrich, and 3 g of 20% [w/v] gum arabic solution from Minerals-Water) was employed as mountant, and samples were placed on glass slides. Samples were observed with an Olympus BX51 light microscope using Nomarski differential interference contrast optics. Photomicrographs were taken with an Olympus DP71 camera (12.5 megapixels) using an Olympus U-TVO.63XC camera adapter, both mounted on the microscope. Images were captured and processed using CELL-B version 2.7 and ImageJ software for stomata, IVG, and vein counting as well as measuring. The areas of stomatal complexes, aperture length, and guard cell width were measured using a Bamboo Pen Tablet from Wacom. Statistical analysis was done using Minitab16 and Graphpad Prism6.
RNA Extraction and Sequencing
For P3 stage leaves, 240 primordia were used per RNA sample; for P4 stage leaves, five primordia were used per sample (P4 stage leaves around 1 cm in length); and for P5 stage leaves, three leaves were used per sample (blade tissue down to the collar only). Samples were harvested between 3 and 5 h into the photoperiod to minimize the potential influence of circadian factors, and the analysis was performed with three biological replicates. RNA was extracted using TriZol (Invitrogen) and cleaned up and DNase1 treated using the Sigma Plant Total RNA kit (Sigma-Aldrich). The quality of the resulting RNA was assessed using the Agilent 2100 BioAnalyzer (www.genomics.agilent.com), and all RNA integrity number values were found to be above 8. RNAseq was carried out at the Liverpool Centre for Genomic Research (www.liv.ac.uk/genomic-research) using RiboZero-treated RNA with library construction following the Illumina TruSeq stranded mRNA protocol (www.illumina.com). Sequencing (Illumina HiSeq 2000) produced paired-end reads with a read length of 100 bp.
Transcript Quantification and Differential Gene Expression Analysis
Paired-end reads were subjected to quality trimming and adaptor filtering using Trimmomatic (Bolger et al., 2014) using the settings LEADING:10, TRAILING:10, SLIDINGWINDOW:5:15, and MINLEN:50. The quality-filtered paired-end reads were then mapped to the complete set of coding sequences from version 7.0 of the japonica rice Michigan State University Release 7 using bowtie2 (Langmead and Salzberg, 2012), and transcript abundances were estimated using RSEM (Li and Dewey, 2011). All pairwise comparisons between developmental stages were made using DESeq (Anders and Huber, 2010), using the default normalization method and identifying differentially expressed genes as those with a Benjamini-Hochberg corrected P ≤ 0.05 (Benjamini and Hochberg, 1995). A principal component analysis plot of all count data by replicate was generated using SIMCA-P+ (version 12).
Validation of RNAseq Data by Quantitative Reverse Transcription-PCR
After RNA analysis by Illumina sequencing, both in silico quality control and quantitative PCR (qPCR) validation of the expression patterns of selected genes were used to assess the quality of the data (Supplemental Fig. S6; Supplemental Table S1). These indicated a low variance between biological replicates and verified that the patterns of expression identified by bioinformatic analysis were reflected at the level of qPCR quantification. Validation of RNAseq data was carried out using RNA retained from the original samples submitted for RNAseq. Moloney murine leukemia virus reverse transcriptase (Invitrogen; www.lifetechnologies.com) was used for complementary DNA (cDNA) synthesis following the manufacturer’s protocol. cDNA from each of three biological replicates for each developmental stage was used, except for P3 stage, where two biological replicates were used. Primers were designed using QuantPrime (Arvidsson et al., 2008; for details, see Supplemental Table S1B). PCR amplification was carried out in an ABI StepOne Plus qPCR system using SYBR Green Master Mix (Invitrogen). Two technical replicates from two separate cDNA synthesis reactions were averaged for each biological replicate, and relative fold changes in expression between developmental stages were calculated using the ƊƊCT method (Schmittgen and Livak, 2008).
Functional Term Enrichment Analysis and Identification of Genes of Interest
The direction of the log₂(fold change) in expression between stages and its Benjamini-Hochberg-corrected significance (P < 0.05) were used to cluster all loci into 11 expression clusters (Supplemental Fig. S7). Custom Perl scripts were used to group genes into clusters. Loci for which there were fewer than 10 fragment alignments were not clustered. The 11 clusters were used to identify the stage at which individual genes reached their maximum expression level.
For MapMan (version 3.5.1R2; Usadel et al., 2005) term enrichment analysis testing, significantly enriched gene groups were identified as those with a Benjamini-Hochberg-corrected P < 0.05 following Wallenius approximation and length normalization of uncorrected P values using GOseq (Young et al., 2010). Up-regulated functional terms were those overrepresented in clusters of genes that were up-regulated between at least two developmental stages and not down-regulated between any two developmental stages. Down-regulated functional terms were those overrepresented in clusters of genes that were down-regulated between at least two developmental stages and not up-regulated between any two developmental stages.
Potential novel regulators of photosynthetic development (Supplemental Table S4A) were identified by applying the following rules: genes must be transcription factors; they must be in cluster up 2 (significantly up-regulated from P3 to P4, no significant change from P4 to P5), up 3 (significantly up-regulated from P3 to P4 and from P4 to P5), or peak (significantly up-regulated from P3 to P4, significantly down-regulated P4 to P5); log₂(fold change) for P3-P4 ≥ 2; level of expression in P4 stage ≥ 5; level of expression in mature leaves ≥ 5 (not for genes in the peak cluster; data from Davidson et al., 2012); and level of expression in rice endosperm ≤ 5 (Davidson et al., 2012).
For genes putatively involved in vascular and stomatal development, genes of interest from Arabidopsis (Arabidopsis thaliana) and maize (Zea mays) were identified from the literature (Scarpella and Meijer, 2004; Liu et al., 2009; Supplemental Tables S5 and S6). Rice orthologs of these genes were identified using conditional orthology assignment as described by Aubry et al. (2014).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number SRP062323.
Results
Photosynthetic Function in Rice Leaves Is Established at the P3/P4 Transition
The first three leaves of a rice plant (P1–P3) are already initiated in the embryo prior to germination. Following germination, additional leaves are produced at regular intervals by the shoot apical meristem (Itoh et al., 2005). Leaf 5 showed a typical and reproducible growth pattern (Fig. 1A) and was chosen for use in all subsequent analyses. Observing developing leaves in rice is complicated by the fact that they are hidden from view by the sheaths of older leaves. Therefore, we characterized a plastochron index (Erickson and Michelini, 1957; Hill and Lord, 1990; Fournier et al., 2005; Fig. 1C), which allowed us to use the length of leaf 3 to robustly assess the developmental stage of leaf 5 without destruction of the plant. Plants could thus be selected and treated at specific plastochron stages of leaf 5 without invasive manipulations. Figure 1, B and D, show the P1 to P6 (mature) stages of leaf 5 growth, and Supplemental Figure S1 provides an overview of the histology of the different stages of rice leaf development.

To investigate at exactly which developmental stage and where within the leaf the ability to generate electron flow via the absorption of light energy occurred, we used chlorophyll fluorescence microscopy. Figure 2 shows a series of example images from leaves at P3, P4, and P5 stages as well as mature leaf blades. Due to their relative size, only portions of the P5 and mature leaf blades are shown. For each sample, the raw fluorescence output is shown adjacent to the calculated values and distribution of φPSII, which provides a measure of the quantum efficiency of electron transport. P3 stage primordia occasionally displayed measurable electron transport (Fig. 2A), but this was very weak and tightly localized to the distal tip of the primordia. Four out of nine P3 primordia analyzed did not display any measurable signal. Early P4 stage leaves showed a much more robust ability to generate electron transport, with all (10 out of 10) samples analyzed showing detectable signal. The signal was again restricted to the distal tip of the leaves but was higher than that observed in P3 samples (Fig. 2B). For comparison, all P5 stage leaves analyzed (which were all visibly green) and all mature leaves analyzed showed a more uniform and very high signal (Fig. 2, C and D).

These physiological data suggested that the ability of rice leaf tissue to perform a basic function of photosynthesis (absorption of light energy to generate an electron flow) develops at the very end of the P3 stage at the distal tip but that there is then a rapid transition to photosynthetic competence during early P4 stage. Analysis of the ultrastructure of plastids during leaf 5 development supported this interpretation. Plastids in P3 primordia lacked any obvious granal structure (Fig. 3A), whereas by P4 stage, plastids with occasional grana were observed (Fig. 3B). In contrast, P5 plastids had distinct grana (Fig. 3C), which looked very similar to those in mature leaves (Fig. 3D). It was noticeable that, even in the P3 stage plastids (i.e. in leaves in which we infer photosynthesis was not occurring or was very limited), distinct starch grains were observed (Fig. 3A). We also investigated the developmental phase of the acquisition of potential photosynthetic capability by examining the pattern of chlorophyll autofluorescence using confocal microscopy. In P4 stage primordia, a clear maximum of signal intensity was observed near the tip of the leaf, with more proximal regions displaying a striated pattern of chlorophyll fluorescence (Fig. 3E).

We did not extract and measure the chlorophyll content of P3 and P4 stage leaves, due to technical difficulties and the oversimplification of the spatial differences in chlorophyll content in P3 and P4 stage leaves this would represent. However, chlorophyll content was around 593 mg m⁻² in mature leaf blades (n = 5; SD = 177 mg m⁻²) and 335 mg m⁻² in P5 stage leaves (n = 5; SD = 54.9 mg m⁻²). The average raw fluorescence signal in regions of P3 and P4 stage leaves where it was detectable was around 10% to 20% (P3 stage) or 10% to 60% (P4 stage) of the raw fluorescence signal observed in mature leaves. Thus, we estimate the average chlorophyll content of these regions of P3 and P4 stage leaves to be around 60 to 120 mg m⁻² (P3 stage leaves) or 60 to 360 mg m⁻² (P4 stage leaves), depending on the sampling location.
To further investigate the nature of the biochemical and physiological events underpinning the very early stages of the acquisition of photosynthetic potential, we analyzed the induction kinetics of φPSII: φPSII = (F ₘ′ − F ₛ)/F ₘ′, where F ₘ′ is maximum PSII fluorescence in the light-adapted state and F ₛ is steady-state fluorescence (Maxwell and Johnson, 2000; Fig. 4, A–D). Data were extracted from up to seven regions of interest (ROIs) along the length of three of the five P3 stage primordia in which φPSII was measurable (Fig. 4A) as well as three P4 stage primordia (Fig. 4B). The results indicated a high degree of variation both along individual primordia (from base to tip) and between primordia, both at the P3 and P4 stages, consistent with the idea that rapid changes were occurring in the capacity for electron transport around this transition. Very fast induction rates were observed in the P3 samples, consistent with P3 leaves being developing sink tissues with high levels of reduced metabolites (Turgeon, 1989; Meng et al., 2001). Slightly slower rates of φPSII induction were observed in tip regions of P4 stage primordia, consistent with our proposal that the generation of Calvin cycle intermediates, the activation of Calvin cycle enzymes, and possibly stomatal opening require more time in this tissue at this stage of leaf development. These data can be compared with those observed in P5 stage and mature leaves, which showed slower induction kinetics but much higher values of φPSII and, overall, more robust patterns of induction despite the higher irradiance used (Fig. 4, C and D). These patterns are characteristic of source leaves in which photosynthetic function has been fully established (Makino et al., 2002).
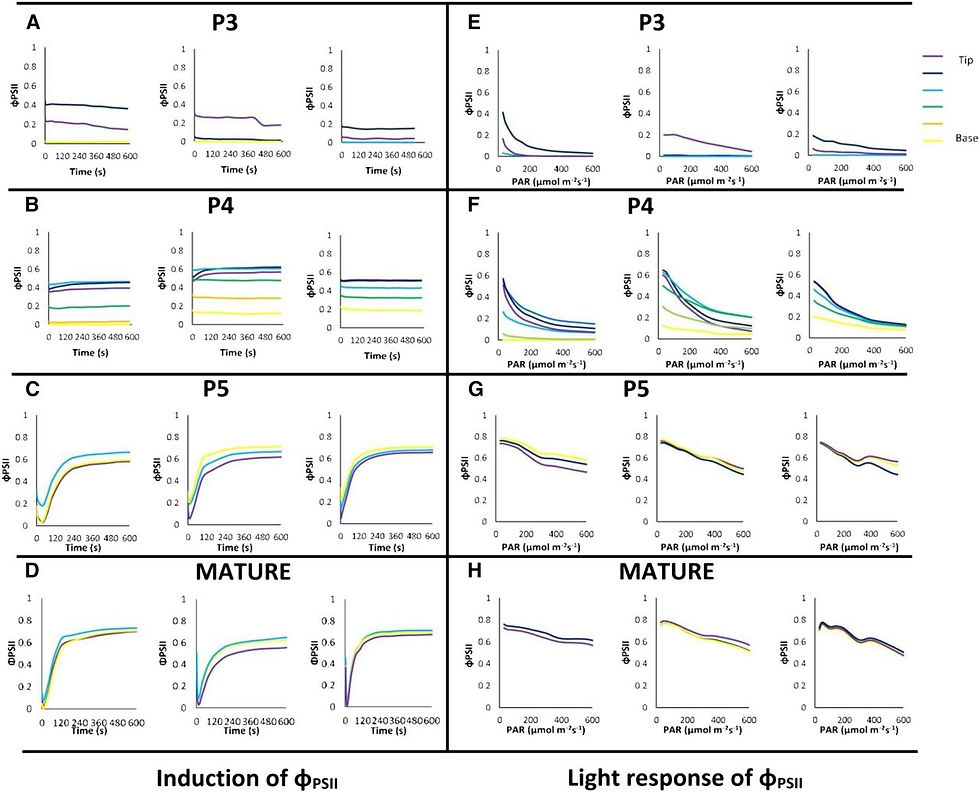
After leaf primordia at different developmental stages had undergone induction and reached a steady state of electron transport, they were exposed to a series of increasing irradiances (Fig. 4, E–H). At each irradiance, steady-state φPSII images were collected and data were extracted from regions along the length of leaf primordia, as described for φPSII induction. In P3 stage leaf primordia, a light response of φPSII was measurable only in the regions nearest the distal tip, and φPSII dropped rapidly to very low levels as the irradiance increased (Fig. 4E). In P4 stage leaf primordia, a light response of φPSII was measurable in regions farther away from the tip than in P3 primordia, and all 10 primordia analyzed still showed a φPSII of 0.2 to 0.1 in regions nearest the tip at the highest irradiance used (Fig. 4F). Generally, the heterogeneity seen in steady-state φPSII values after induction was also seen in the light response of φPSII in P3 stage leaves (Fig. 4, A and E). This can be compared with the pattern of light induction seen in P5 and mature leaf samples (Fig. 4, G and H), which showed relatively consistent induction kinetics along the length of the leaves analyzed and high consistency between different samples. Nonphotosynthetic quenching was also measured during induction and at a range of irradiances, but slight movement of the samples between recording of the F ₘ and F ₘ′ in P3 and P4 stage leaves limited the interpretation of these data (Supplemental Fig. S3).
A Loss of Leaf Developmental Plasticity Occurs after the P3 Stage
The above data highlighted the P3/P4 transition as a key stage in the transition to photosynthetic competence during rice leaf development. To investigate to what extent this physiological transition correlated with specific elements of leaf differentiation, we performed a histological analysis of leaf 5 from P2 through P6. At P2 stage, there was little overt cellular differentiation to indicate the future position of vascular tissue across the leaf blade (Fig. 5A; Supplemental Fig. S1). By P3 stage, coordinated patterns of cell division indicated regions of vascular formation that became more pronounced during P4, so that by P5 stage and the mature P6 stage, a classical pattern of vascular differentiation into xylem and phloem was apparent (Fig. 5A).
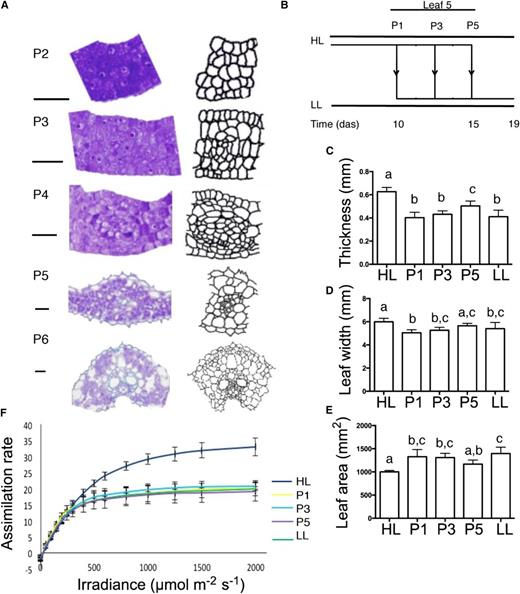
It has long been established that rice leaves grown under different irradiances undergo a process of acclimation to yield distinct leaf morphologies (Murchie et al., 2005). To investigate the degree of developmental plasticity in this system and to define when this plasticity is lost, we performed a series of experiments in which plants containing leaf 5 at particular developmental stages (P1, P3, and P5) were transferred from a high-light (HL; 700 µmol m⁻² s⁻¹) to a low-light (LL; 250 µmol m⁻² s⁻¹) environment and the structure and photosynthetic function of the mature leaves were measured (Fig. 5, B–F). As shown in Figure 5C, leaf 5 grown continuously under an HL environment had a mean thickness of 0.63 mm, whereas leaf 5 grown continuously under an LL environment was significantly thinner (0.41 mm; P < 0.05, Tukey’s honestly significant difference test). When plants were transferred from HL to LL conditions when leaf 5 was at P1 or P3 stage, the mature leaves attained an appropriate thickness for the new LL environment (0.4 mm when transferred at P1 stage and 0.43 mm when transferred at P3 stage). However, when the transfer occurred at the P5 stage, the mature leaf was of intermediate thickness (0.5 mm), indicating that at the P5 stage, leaves were no longer able to morphologically acclimate fully to the new light environment, consistent with previous work (Murchie et al., 2005). Measurement of leaf blade width (Fig. 5D) and area (Fig. 5E) revealed that growth in the surface plane of the blade showed a similar pattern of determination by the P5 stage with respect to light environment as leaf thickness.
Combined gas-exchange/chlorophyll fluorescence analysis indicated that leaf 5 of plants grown under continual HL conditions had a higher assimilation rate at ambient CO2 than the equivalent leaf grown under LL conditions (Fig. 5F). When leaf 5 was transferred from HL to LL conditions at the P1, P3, or P5 stage, it was able to physiologically acclimate at all stages so that the measured leaf assimilation rate in the mature leaf was similar to a leaf maintained continuously under LL conditions. These data indicated that photosynthetic capacity could acclimate post-P5 via altered biochemistry, consistent with previous observations (Murchie et al., 2002, 2005).
Having established that rice leaves were showing an appropriate acclimation response at the gross morphological level, we set out to investigate the light response at the cellular level, with particular emphasis on stomatal patterning, since one might expect this to relate to the acquisition of photosynthetic function. Stomata in rice leaves form in specific epidermal cell files (Fig. 6A; Luo et al., 2012). Epidermal cell files are formed in a series of IVGs defined by parallel vascular strands along the long axis of the leaf. SD within each IVG at the midpoint along mature leaf 5 was measured in plants grown under HL and LL conditions (Fig. 6B). Under HL conditions, leaves had a maximum of 16 IVGs, whereas under LL conditions, up to 18 IVGs formed. Values of SD were found to be reasonably constant across all IVGs under both treatments, with the exception of the IVGs at the leaf margin, which tended to have a relatively high SD. With respect to IVD, these were generally smaller in LL conditions across all regions of the leaf (Fig. 6C). Again, the IVD values at the leaf margin for both HL and LL leaves tended to be different from the rest, with the margin IVD being relatively small, mirroring the relatively high SD values observed in this IVG (Fig. 6B).

As in other grass leaves, stomata in rice are formed via an intricate but stereotypical series of cell divisions leading to the development of a stomatal complex consisting of a pair of guard cells and supporting cells (Fig. 6E; Luo et al., 2012). When SCA was measured in mature HL-grown leaves, a relatively consistent value was observed in all IVGs (Fig. 6D). When leaves were grown under continual LL conditions, a much smaller SCA was measured in all IVGs (Fig. 6D). To further investigate the nature of this difference in SCA and to identify the developmental window during which SCA was set, we analyzed stomata in a series of plants that had been transferred from HL to LL conditions at specific stages of development of leaf 5 (as described in Fig. 5B). These data displayed a startling significant difference in variance between samples transferred at different developmental stages (Brown-Forsythe test, P < 0.001), with leaves transferred at P1 and P3 stages showing very high variances (Fig. 6F). A nonparametric analysis of the data (Kruskal-Wallis test with posthoc Dunn’s multiple comparison) indicated that the median SCA values for P1-transferred and LL samples were significantly different from the other treatments (P < 0.05), suggesting a fluidity in the setting of stomatal size in early rice leaf development that is later lost.
To investigate the cause of this variation in SCA, we measured SCL and SCW. A similar pattern was observed to that recorded for SCA, suggesting that the variation observed in SCA reflected changes in both SCL and SCW (Fig. 6, G and H). Comparison of the different SCL samples revealed a significant difference in variance (Brown-Forsythe test, P < 0.01), with P1- and P3-transferred samples showing the highest variances, but a nonparametric test did not distinguish any of the samples for SCL. The SCW data also showed a significant variation in variance (Brown-Forsythe test, P < 0.05), and the median values obtained from leaves transferred at P1 and P3 stages could be distinguished from the other treatments (Kruskal-Wallis test with Dunn’s multiple comparison, P < 0.05).
Interestingly, although comparison of the mean values of SD in the transferred leaves indicated no difference between the different treatments, the data indicated a higher variance in SD in the P1- and P3-transferred leaves compared with the other samples (Fig. 6I), with some transferred leaves showing very low (fewer than 150 stomata mm⁻²) or very high (more than 600 stomata mm⁻²) values (Supplemental Fig. S4). Thus, a plasticity in the setting of various stomatal parameters was observed during very early leaf development that seemed to be lost later.
An analysis of stomatal differentiation by scanning electron microscopy supported the interpretation that early stage primordia (P1 and P3) show greatest flexibility in the light response of SCA. At P3 stage, epidermal cell files were clearly visible along the surface of the primordium, but no overt signs of stomatal differentiation were apparent (Fig. 6, N and O). By late P4 stage, differentiated stomata were clearly visible (Fig. 6P), consistent with the P4 stage being a phase of rapid stomatal differentiation, subsequent to patterning events during P3.
Since stomata in rice form along epidermal cell files, we also investigated the extent to which the parameters of stomatal size and density might be set by the overall parameters of epidermal CFN and CFW within IVGs and the relationship of these parameters to whole-leaf values of vein number and IVD. The overall pattern of CFW was similar to that observed for SCA (Fig. 6, F and J). The differences in variance for CFW were less than for SCA, and the P1- and P3-transferred samples had a significantly lower CFW than the other treatments (ANOVA with posthoc Tukey’s honestly significant difference test, P < 0.05).
The differences in CFW were accompanied by differences in IVD (Fig. 6K). Most notably, the P1-transferred leaves had a significantly smaller IVD compared with HL leaves (P < 0.005), and LL leaves also had smaller average IVD (P < 0.01). A decrease in IVD is predicted to lead to an increase in the frequency of veins across the leaf blade (since veins set the boundaries for IVD). This was observed for the P1-transferred leaves, which showed a significantly higher vein frequency than the other treatments (P < 0.05; Fig. 5L). It is interesting that, although under HL conditions IVD was higher than under LL conditions and CFN was essentially unchanged (Fig. 6M), vein frequency across the blade of HL and LL leaves was very similar (Fig. 6L). This apparent discrepancy was resolved by the observation that, under HL conditions, the width of the vascular bundles was smaller than under LL conditions. Thus, there appeared to be a compensation process between IVD and vascular bundle width that acted to maintain vein frequency and a relatively constant number of intervening epidermal cell files.
In addition to investigating the outcome on leaf development of transitions from HL to LL conditions, we also analyzed the reverse situation, where leaf primordia at specific plastochron stages from plants grown under LL conditions were transferred to HL conditions. For this analysis, we focused on those traits that showed the most marked differences in the high- to low-irradiance experiments described above. As shown in Supplemental Figure S5A, leaves grown under LL conditions and then transferred to HL conditions at P5 stage did not attain a typical HL thickness at maturity, whereas leaves transferred at the P3 stage achieved a thickness that could not be discriminated from leaves continually kept under HL conditions. Interestingly, P1 stage leaves transferred to HL conditions remained relatively thin at maturity, suggesting that, under some conditions, very early stage primordia can remain fixed with respect to final leaf thickness. When SCA was analyzed, a similar pattern was observed, with the P5-transferred leaves displaying a mean SCA that was similar to that observed under LL conditions and distinct from that observed under HL conditions (Supplemental Fig. S5B), with P1- and P3-transferred leaves showing a more intermediate phenotype. There was no difference in mean SD between the different treatments (Supplemental Fig. S5C), but there was a trend for higher variance in the leaves transferred at P3 and P5 stages, with densities of over 500 stomata mm⁻² being observed. Similarly, the various treatments did not lead to a significant difference in final CFW, although there was a trend again for higher variance in leaves transferred at P3 and P5 stages (Supplemental Fig. S5D).
Gene Expression Patterns Underpinning the P3/P4 Transition
Our analysis of both physiology and structure identified the P3/P4 transition as an important stage in rice leaf development with respect to the acquisition of photosynthetic capability and associated cellular differentiation, as well as the ability to respond to environmental signals by altering aspects of cellular differentiation. To investigate the gene expression changes underpinning this transition, we performed RNA sequencing (RNAseq) analysis on leaf 5 at P3 stage and P4 stage and on the leaf blade at P5 stage (Supplemental Data S1). Overall, 14,502 genes were identified that were differentially expressed during early leaf development (out of a total of 25,768 for which expression was detected). Having validated the RNAseq data (Supplemental Fig. S6), we interrogated the data sets to identify patterns of gene expression during early leaf development that might underpin the key processes identified by our morphological and physiological analysis. These results are summarized in Figure 7 and described below.

Since our analysis indicated that photosynthetic electron transport first develops at the P3/P4 stage transition, we first investigated the pattern of transcript accumulation for those genes annotated as photosynthetic (MapMan bin PS).
Our results indicated that 40 genes involved in photosynthesis (out of 351 rice genes annotated with MapMan bin PS) were already at 50% or more of their mature leaf expression level at the earliest developmental stage studied, P3 (Supplemental Table S2), suggesting that a small number of genes involved in the photosynthetic process are already transcriptionally expressed in tissues that are not yet photosynthetically active. The most common expression pattern of genes annotated as involved in photosynthesis was an increase in expression from P3 to P4 and from P4 to P5 stages (159 genes; Supplemental Table S3A). Broadly, functional groups of genes up-regulated from P3 to P5 stage were found to include those involved in the Calvin-Benson cycle, photosynthetic light reactions, and subunits of both PSI and PSII. These expression changes were accompanied by the up-regulation of photoprotective mechanisms such as carotenoid biosynthesis (18 of 21 annotated genes; Supplemental Table S3B) and around half of genes involved in ascorbate and glutathione metabolism (49 of 106 annotated genes; Supplemental Table S3C).
A range of expression patterns was seen in genes identified previously through rice mutant studies as playing a role in photosynthetic development (Table I). A small number of these were most highly expressed at P3 stage. However, many other genes with known rice photosynthetic mutant phenotypes showed a peak in expression at P4 stage, as well as several genes with Arabidopsis (Arabidopsis thaliana) photosynthetic mutant phenotypes. A further group of genes with rice photosynthetic mutant phenotypes were maximally expressed at P5 stage. Genes showing this expression pattern also included orthologs of those known to play a role in photosystem assembly in Arabidopsis. Only a small number of genes that have known mutant phenotypes involved with photosynthesis in rice showed a neutral expression pattern in these early stages of leaf development.
Table I. Expression patterns of rice genes known to be involved in photosynthetic development
Locus | Gene Name(s) | Reference | Cluster |
LOC_Os09g38980 | TCD9 | Jiang et al. (2014b) | down 1 |
LOC_Os05g37160 | OsFtsz2 | Vitha et al. (2001) | down 1 |
LOC_Os06g07210 | RNRL1 | Yoo et al. (2009) | down 3 |
LOC_Os06g14620 | RNRS1 | Yoo et al. (2009) | down 3 |
LOC_Os02g56100 | RNRL2 | Yoo et al. (2009) | down 3 |
LOC_Os06g03720 | RNRS2 | Yoo et al. (2009) | down 3 |
LOC_Os08g07840 | OsPOLP1 | Takeuchi et al. (2007) | down 3 |
LOC_Os03g20460 | OsGKpm, VIRESCENT2 | Sugimoto et al. (2007) | neutral |
LOC_Os03g16430 | SIG2B | Kasai et al. (2004) | neutral |
LOC_Os02g39340 | OsDG2 | Jiang et al. (2014a) | neutral |
LOC_Os03g45400 | NUS1, VIRESCENT1 | Kusumi et al. (1997); Kusumi et al. (2011) | peak |
LOC_Osp1g00240 | rpoB | Hanaoka et al. (2005) | peak |
LOC_Osp1g00250 | rpoC1 | Hanaoka et al. (2005) | peak |
LOC_Osp1g00260 | rpoC2 | Hanaoka et al. (2005) | peak |
LOC_Os11g26160 | SIG2A | Kasai et al. (2004) | peak |
LOC_Os06g44230 | OsRpoTp | Kusumi et al. (2004) | peak |
LOC_Os09g21250 | YLC1 | Zhou et al. (2013) | peak |
LOC_Os05g23740 | OsHsp70CP1 | Kim and An (2013) | peak |
LOC_Os03g29810 | VYL, OsClp6 | Dong et al. (2013) | peak |
LOC_Os05g32680 | PAC | Meurer et al. (1998) | peak |
LOC_Os04g56970 | OsFtsz1 | Vitha et al. (2001) | peak |
LOC_Os04g42030 | SNOW-WHITE LEAF1 | Hayashi-Tsugane et al. (2014) | peak |
LOC_Os08g06630 | SIG1 | Kasai et al. (2004) | up 1 |
LOC_Osp1g00660 | rpoA | Hanaoka et al. (2005) | up 2 |
LOC_Os05g51150 | SIG3 | Kasai et al. (2004) | up 2 |
LOC_Os04g39970 | OsV4 | Gong et al. (2014) | up 2 |
LOC_Os05g50930 | SIG5 | Kasai et al. (2004) | up 3 |
LOC_Os08g14450 | SIG6 | Kasai et al. (2004) | up 3 |
LOC_Os06g40080 | YGL2 | Chen et al. (2013) | up 3 |
LOC_Os10g35370 | PORB | Kang et al. (2015) | up 3 |
Not found in rice | SIG4 | Kasai et al. (2004) | Not applicable |
To investigate the control of expression of these photosynthesis-associated genes at the P3/P4 stages, we also analyzed transcription factors showing similar patterns to these known photosynthetic genes during rice leaf development, which led to the identification of 30 genes with increasing expression patterns during early leaf development and 21 genes whose transcripts peaked at P4 stage (for full selection criteria, see “Materials and Methods”; Supplemental Table S4A). This list included genes shown previously to play a role in regulating chloroplast development (e.g. GLK1 and GLK2; Fitter et al., 2002; Wang et al., 2013a), supporting the notion that the other transcription factor genes in this list may also play a role in facilitating or regulating the onset of photosynthesis. Notably, there are also several CCT family transcription factors in our list of potential regulators of photosynthetic development.
The setting of aspects of CFW, SCA, and vein frequency, as well as the overall parameters of leaf width and thickness, prior to the P5 stage suggested that genes involved in cell growth and division at the P3/P4 transition play an important role in determining morphological parameters linked to photosynthesis and acclimation. In line with this, functional groups of genes down-regulated from P3 to P5 stage included those involved in the cell cycle (24 genes, including ARF75, TANGLED1, and many cyclins), regulation of transcription (270 genes, including transcription factors and chromatin-remodeling factors), DNA synthesis (97 genes, including histones and DNA polymerase subunits), and protein synthesis (both plastid and nonplastid; 108 genes, mostly involved in ribosome biogenesis). Notably, the P3/P4 transition was associated with an increase in the expression of genes associated with the inhibition of progress through the cell cycle (KRP1 and KRP6) and an increased relative expression of genes associated with cell wall differentiation and remodeling.
The P3/P4 transition is associated with vascular differentiation and patterning (Fig. 5A), and of the 36 rice genes identified as vascular development regulators or orthologs of vascular development regulators in Arabidopsis (Scarpella and Meijer, 2004; Supplemental Table S5), 22 reached their highest level of expression by P3 stage, nine peaked at P4 stage, and only five were expressed most highly at P5 stage (Fig. 7B). Thus, most of these putative vascular development regulators showed a decrease in expression from P3 to P5. Of the few genes known to have a vascular mutant phenotype in rice, several were most highly expressed at P3 stage, including OsKn3, OsAGO7, and NARROW AND ROLLED LEAF1 (Postma-Haarsma et al., 1999; Shi et al., 2007; Hu et al., 2010). However, a neutral expression pattern across the three stages studied was observed for the NARROW LEAF1, NARROW LEAF2, and NARROW LEAF3 genes (Qi et al., 2008; Cho et al., 2013). As auxin has long been known to be involved in vascular development, we also studied the expression patterns of OsPIN genes found previously to be expressed in the vasculature (Wang et al., 2009). These genes showed a mixture of expression patterns, with OsPIN1a most highly expressed at P3 stage, OsPIN5b showing a neutral expression pattern, OsPIN10a peaking at P4 stage, and OsPIN5a and OsPIN1b reaching maximal expression at P5 stage. The expression of OsPIN2 was not detected.
Finally, since our results indicated that the P3 stage was characterized by a plastic response in terms of stomatal size in response to altered light environment (Fig. 6) but that this plasticity was lost by P5 stage, we interrogated our data to identify which rice genes and orthologs of maize and Arabidopsis genes linked with stomatal differentiation and patterning were expressed during P3 and P4 stages. Of the 17 genes identified as putative stomatal development regulators in rice, 11 were most highly expressed at P3 stage (Fig. 7B; Supplemental Table S6). This included orthologs of SPEECHLESS, TOO MANY MOUTHS, EPF2, EPFL7, and YODA (Bergmann et al., 2004; Liu et al., 2009; Rychel et al., 2010). A further five reached their highest expression level at P4 stage, including rice orthologs of SCREAM1, EPFL9, and MUTE (Liu et al., 2009; Hunt et al., 2010). The expression of all stomatal differentiation genes was less in P5 than in earlier stages.
Discussion
Despite the importance of leaves as the basic engine for photosynthesis-derived nutrition, aspects of our understanding of how these organs develop in monocots are incomplete. In this article, we set out to address this knowledge gap by addressing some specific questions. When does a rice leaf gain capacity for light capture to generate reducing power for carbon fixation? How well does this acquisition of physiological capability correlate with morphological aspects of leaf differentiation and with underlying patterns of gene expression? How flexible are these developmental processes, and when is developmental plasticity lost?
The P3/P4 Transition Is When Photosynthetic Competence Is Initiated
By creating a plastochron index, we were able to stage rice leaf development in a robust fashion, enabling comparison of equivalent leaves. Although the main structural and anatomical changes that occur during rice leaf development have been characterized (Itoh et al., 2005), our results provide an analysis of how this structure relates to function (i.e. when a leaf actually gains the capacity to perform an essential element of photosynthesis, such as absorption of light energy to generate an electron flow).
Although the microfluorescence technique used for this analysis places some constraints on data interpretation (e.g. no absolute measures of electron transport rate or photosynthetic capacity were calculated, since absorbance was found to be variable [Supplemental Fig. S2], and other assumptions made in the comparison of these very different tissues may not be met), we are confident that true quenching of chlorophyll fluorescence was occurring in our primordia and that our φPSII measurements are valid. We cannot rule out the presence of some cyclic electron flow, reduction of the plastoquinone pool in the dark (Corneille et al., 1998), or the presence of a small proportion of light-harvesting complexes not connected to reaction centers. However, we did not observe the very high raw fluorescence signal that might be observed if a large proportion of light-harvesting complexes were uncoupled and the reasonable Fᵥ/Fₘ (maximum photochemical efficiency of PSII in the dark-adapted state) values observed indicate nonzero amounts of quenching. Furthermore, the light response of φPSII observed in primordia shows the expected pattern of decreasing φPSII at increasing irradiances, consistent with a functioning electron transport chain.
Our results indicate that the ability to absorb light energy to generate an electron flow is gained at the very end of the P3 stage of development at the distal tip of the primordium, followed by a very rapid transition to photosynthetic competence during early P4. Electron transport efficiency does not reach the level of that of a mature leaf until the P5 stage, while little additional photosynthetic electron transport efficiency is gained between P5 stage and the mature leaf. The high variability of the light response of φPSII between samples and between regions is consistent with P3 and P4 stage primordia undergoing rapid photosynthetic development, with different leaves having widely differing abilities to use increasing amounts of light to drive electron transport. This interpretation was supported by a structural analysis of plastid differentiation. Plastids prior to P3 stage lacked obvious grana, whereas by P4 stage, plastids had internal structure consistent with a capacity for photosynthetic activity. P3 plastids frequently contained starch granules, suggesting that the plastids were importing carbon from source leaves and that starch grains were being used as a temporary carbon store, potentially enabling rapid growth during the fundamental biochemical switch from sink to source metabolism. Recently, Kusumi et al. (2010) used a similar structural analysis of plastid differentiation alongside chlorophyll fluorescence imaging to show that, in rice, leaves at the P4-2 stage (around 2 cm long; before emergence) have measureable φPSII. Other than that of Kusumi et al. (2010), there are few studies reporting photosynthetic measurements on very young leaves. Meng et al. (2001) showed that, in the developmental gradient present in a developing tobacco (Nicotiana tabacum) leaf (the youngest used was 3.9 cm long), even regions at the base have a measurable rate of electron transport. Peterson et al. (2014) reported changes in the relative abundance of PSI and PSII along maize leaves (the third leaf of 12-d-old seedlings was used) and concluded that redox mediation of chlorophyll biosynthesis may regulate photosystem assembly and thus ensure the development of a suitable PSI/PSII excitation balance. In the most immature (basal) leaf segment studied, Peterson et al. (2014) also reported a larger nonvariable component of fluorescence, which they attribute to a moderate accumulation of incompletely assembled, nonfunctional PSII complexes. However, no detailed measurements of φPSII kinetics at different developmental stages of the leaf were reported in those studies. Our data agree with those of Kusumi et al. (2010), but we also show that the first detectable electron transport occurs before this in the tip of P3 stage leaves and that the development of photosynthetic function occurs basipetally thereafter. Thus, the large longitudinal developmental gradient in the acquisition of φPSII in P3 and P4 stage primordia can be visualized to a high spatial resolution using our approach.
These physiological data can be compared with our mRNA analysis. In particular, we addressed two questions. Which photosynthetic genes become transcriptionally active at the onset of the physiological process? Is there a core set of early photosynthetic genes in rice?
Analysis of gene expression in the very earliest stage of leaf initiation in maize indicated that there is no significant difference in the expression of photosynthesis-related genes between the shoot apical meristem proper and leaves at the P0/P1 stages (Brooks et al., 2009), whereas in tomato (Solanum lycopersicum; a dicot), a gene encoding the small subunit of Rubisco was already expressed at P1 stage but not expressed in the shoot apical meristem (Fleming et al., 1993). In another study, it was found that, in older intact maize seedlings, 39 photosynthesis-related genes were up-regulated compared with shoot apical meristems (Ohtsu et al., 2007). However, both studies in maize used microarrays enriched in probes reflecting specific subsets of tissues, limiting the extent to which the results can be compared with our RNAseq-based analysis (Ohtsu et al., 2007; Brooks et al., 2009). In rice, we found that, by the P3 stage, 40 genes involved in photosynthesis (out of 351 rice genes annotated as photosynthetic) were already at 50% or more of their mature leaf expression levels. These data suggest that over one-tenth of genes involved in photosynthesis are already highly expressed before a leaf can actually utilize the encoded enzymatic machinery. Since light is required for the final step in chlorophyll biosynthesis, and our analysis indicated a wave of acquisition of chlorophyll fluorescence from the primordium tip toward the base at the P3/P4 transition, our data suggest that the basic machinery for photosynthetic electron transport is developmentally set prior to the late P3 stage by a core set of genes, with a light-related trigger initiating the actual capability for photosynthetic electron flow. Analysis of P2 stage primordia would allow further testing of this hypothesis; however, we found the dissection of such young primordia in rice to be technically very challenging.
Many of the genes that are maximally expressed at P4 stage are known to have chloroplast biogenesis mutant phenotypes, emphasizing the importance of this stage for the development of photosynthetic capability. Transcripts for most genes encoding the core photosynthetic machinery (including those for most electron transport chain components and Calvin-Benson cycle enzymes) increase from both P3 to P4 and P4 to P5, showing that bulk production of these vital components of autotrophy occurs over a relatively longer developmental time. Kusumi et al. (2010) split photosynthetic development in rice into three stages: DNA replication and plastid division (late P3/early P4 stage); activation of the chloroplast genetic system (particularly the transcription/translation machinery; early P4 stage); and activation of the photosynthetic machinery (late P4 stage to P5 stage and beyond). Our results are largely consistent with their findings. However, we also pinpoint a varied set of photosynthesis-related genes that are already expressed at the very early P3 stage, we add more genes with known mutant phenotypes into the expression picture, and we identify a novel set of transcription factors that show expression changes accompanying those of these known genes. This diverse list of transcription factors contains known regulators of photosynthetic development as well as many CCT family genes. In Arabidopsis, the CCT family gene CIA2 was found to regulate chloroplast protein synthesis and import (Sun et al., 2009). Thus, this list of transcription factors may provide a useful toolbox for future modification of photosynthetic function in rice.
The developmental stages in which we have studied the transcriptomic dynamics are directly comparable to those studied previously in maize (Wang et al., 2013b). In these maize primordia, key photosynthetic genes, such as those encoding Calvin cycle enzymes and PSII reaction center subunits, are already expressed at low levels and are up-regulated in later developmental stages. In maize primordia, the chlorophyll biosynthetic enzyme PORB seems not to be expressed until later developmental stages (equivalent to rice P4/P5), which is also when tetrapyrrole biosynthesis shows a peak of activity. Other available data sets on monocot developmental transcriptomics are from segments along the maize leaf developmental gradient (Li et al., 2010; Tausta et al., 2014; the third leaf of 9-d-old seedlings was used). Although it is more difficult to directly compare these studies of leaf segments with our entire (but much younger) primordia, the general pattern of photosynthetic genes already being expressed in the youngest segments at the base of the leaf (before the sink-source transition) is consistent with our findings.
Major Patterning Events Linked to Photosynthesis Occur in the Leaf at the P3/P4 Transition
In addition to the appropriate biochemical apparatus for light capture and carbon fixation, photosynthesis requires the integrated supply and export of material, including CO2, water, carbon, and nitrogen. Since our physiological analysis indicated that the P3/P4 transition is key to the acquisition of photosynthetic competence, we investigated whether associated elements of leaf differentiation, including vasculature and stomata, were coordinated with this phase. Our data indicate that this is the case and identify a series of genes whose expression is likely to be involved in the appropriate spatial and temporal differentiation of these elements. Undoubtedly, the development of morphological characteristics is affected by the longitudinal developmental gradient existing within rice leaf primordia, as visualized in our chlorophyll fluorescence imaging analysis. We captured precisely positioned snapshots along this gradient that demonstrate major changes in morphological development at the P3, P4, and P5 developmental stages. This provides guidance for the interpretation of our stage-specific gene expression data.
Stomatal differentiation occurs during P4 stage, and our analysis revealed the expression during this phase of a number of genes associated in other systems with processes of stomatal patterning and differentiation. Previous attempts to detect the expression of some of these genes in rice (e.g. OsSPCH1 and OsSPCH2) have failed (Liu et al., 2009), suggesting that the highly targeted and staged RNAseq approach taken here was essential for the detection of these transcripts. The overall pattern of stomata-associated gene expression was consistent with the observed limitation of stomatal differentiation to the P3/P4 stage. Expression of most of these genes was minimal in P5 stage leaves, and the others tended to show a gradual decline in expression from P3 to P5, with some showing a peak in expression at P4 (e.g. OsMUTE, OsSCREAM1, and OsEPFL9). In light of the role of these genes in controlling stomatal patterning and differentiation in other systems, these rice genes likely play a vital role in setting parameters of stomatal pattern and number.
Since there is significant interest in modifying aspects of rice leaf structure for crop improvement, we thought it would be informative to explore the limits of flexibility of the normal process of leaf differentiation, since this would both inform on the boundaries of the system and identify whether particular stages of development should be targeted for modification. To do this, we exploited the knowledge that transfer of rice plants from high to low irradiance leads to an acclimation process in developing leaves so that morphology appropriate to the new light environment is generated. Focusing on stomata, these experiments supported and extended previous observations that this morphological acclimation process occurs prior to the P5 stage, whereas photosynthetic capacity can acclimate post-P5 stage through altered biochemistry (Murchie et al., 2005). Our data indicate that stomata in high-irradiance-grown rice are larger than those grown under low irradiance but with no significant shift in mean stomatal density. Most strikingly, they indicate that at the P1 and P3 stages, the process of stomatal patterning is highly plastic and can respond to a shift in irradiance to generate mature leaves with high variation in stomatal density and size. Thus, rice leaves do possess an inherent plasticity in the stomatal patterning mechanism that is subject to environmental regulation, but this is limited to the P1 to P3 stages. This is clearest in leaves transferred from HL to LL conditions but is also apparent in leaves transferred in a reciprocal fashion from LL to HL conditions. The situation here is slightly more complex, but again, it is the P3 stage that appears to be the key developmental phase in terms of the plasticity of response to the external environment. Although well documented in eudicots, the mechanism of systemic control of leaf development by environmental factors remains unclear (Lake et al., 2001, 2002; Murchie et al., 2005; Coupe et al., 2006). Our results suggest that manipulation of this system could allow the generation of rice leaves with very distinct stomatal properties and, thus, water relations. We suggest that targeting the P3/P4 stage of development would be most appropriate for such manipulations.
Due to the anatomy of rice leaves, the width of a stomatal complex will be influenced by the width of the epidermal cell file within which it arises. Coupled to the number of cell files within an IVG and the number of IVGs in a leaf (which will be set by the number of veins and the overall width of a leaf), a complex interaction of cellular and leaf-scale patterning and differentiation events will influence the final number and size of stomata. Our data show that transfer of a rice leaf from an HL to an LL environment at a very early stage of development (P1) leads to narrower leaves with an increased number of (but smaller) IVGs. The absolute number of cell files within these narrower IVGs remains relatively constant, but the width of the cell files tends to be smaller (i.e. the leaf acclimates to the new environment at the cellular level). At the level of gene expression, genes involved in cell division processes tend to decrease in transcript level from P3 through P5, but it is interesting that some members of the KRP gene family (associated with the termination of cell division; De Veylder et al., 2001; Barrôco et al., 2006) show peaks of relative transcript level at the P3 and P5 stages. Modulation of expression of these genes may play a role in determining the number and size of cells as leaves enter the P5 stage of rice leaf maturation.
Our data indicate that changes in epidermal cell file number and size were linked with changes in vein number and size. As in maize, the midvein is initiated early (by rice P2 stage, maize P1/P2 stage), with the development of lateral veins occurring at the next developmental stage (rice P3 stage, maize P3/P4 stages; Wang et al., 2013a). However, in maize, foliar leaves develop the spacing (by P4 stage) and characteristic Kranz anatomy (by P5 stage) of C4 plants, which does not occur in our C3 rice leaves. The venation pattern that develops in rice is instead broadly similar to that of many other C3 grasses (Sakaguchi and Fukuda, 2008; Sage and Sage, 2009; for review, see Nelson and Dengler, 1997).
To investigate the regulation of vascular development in rice further, we studied the developmental timing of expression in rice of a number of genes previously linked with the control of vascular development in rice, Arabidopsis, and maize (Scarpella and Meijer, 2004). Most of these genes reached maximal expression at the P3 stage followed by a general decrease by P5. The general decline in the number of highly expressed vascular-linked genes from P3 to P5 coincides with the decrease in the rate of differentiation seen at these later stages, suggesting that this loss of gene expression may limit the ability of older stages to initiate new vascular tissue. Mutation in some of these genes has been shown to lead to altered vascular differentiation in rice (Nishimura et al., 2002; Yamaguchi et al., 2004), suggesting that they represent a panel of relevance for the future investigation of rice leaf vascular development. However, the number of rice vein patterning mutants described at the molecular level is very limited, and many of those mentioned here show pleiotropic developmental defects, including a strong link between vein patterning and leaf width (Qi et al., 2008). Thus, suitable targets for the manipulation of rice vascular patterning remain elusive.
How the differentiation of vascular and surrounding photosynthetic tissue is organized has been much debated (Berleth et al., 2000; Scarpella et al., 2003). Chlorophyll fluorescence arose in stripes along the developing leaf, consistent with the idea that, already by the early P4 stage, leaf tissue was set on a developmental trajectory setting apart nonphotosynthetic vascular and photosynthetic intervening mesophyll tissue. The differentiation of photosynthetic tissue may be organized in reference to the network of developing vascular tissue, which provides a scaffold from which positional signals inform the differentiation of the mesophyll. Conversely, Andriankaja et al. (2012) showed that the exit from proliferation of Arabidopsis leaf cells may be regulated by their photosynthetic differentiation, and Scarpella et al. (2004) demonstrated in Arabidopsis that mesophyll differentiation terminates the iterative process of vascular initiation. Which of these processes is at play in monocots, and rice specifically, is unclear. However, we provide evidence that vascular development and the initiation of photosynthetic differentation are temporally and spatially coordinated in rice, with both occurring around the P3/P4 stages. In addition, the striated pattern of chlorophyll autofluorescence observed in P4 stage primordia (Fig. 3) is reminiscent of vascular patterning. Direct evidence for positional signals from the vasculature informing photosynthetic development in rice is not available; however, in plants with very clear compartmentalization of photosynthetic and nonphotosynthetic function, such as the C4 plant maize, such positional information is known to be key (Nelson and Langdale, 1989). The molecular nature of positional information derived from the vasculature has begun to be pinned down in root development (Sabatini et al., 1999; Nakajima et al., 2001; Wu et al., 2014), suggesting that this is a tractable problem.
Conclusion
The results reported here identify the P3/P4 transition as a highly dynamic stage in rice leaf development where initial competence for photosynthesis is achieved. This is coordinated with processes of vascular and stomatal differentiation, and we identify sets of genes involved in the setting of each of these processes. As well as identifying targets for future manipulation, our results suggest that such manipulations should target stages prior to P4 to exploit the endogenous plasticity of differentiation exhibited by rice leaves during this phase of development.
Supplemental Data
Supplemental Figures








Supplemental Tables
Supplemental Table S1. Verification of RNAseq results by qPCR and qPCR primer details.
Supplemental Table S2. Genes annotated with MapMan bin PS that reach 50% or greater maximal expression by P3 stage.
Supplemental Table S3. Genes annotated with MapMan bins PS, Carotenoids, or Ascorbate and Glutathione and their expression patterns during rice early leaf development.
Supplemental Table S4. Transcription factors that are potential regulators of photosynthetic development.
Supplemental Table S5. Arabidopsis and maize genes known to regulate vascular development and their putative rice orthologs, and expression patterns of other rice genes known to be involved in vascular development.
Supplemental Table S6. Putative regulators of stomatal development in rice and their expression patterns during early leaf development.
Supplemental Data S1. All rice RNAseq count data, median normalized.
Glossary
φPSII
quantum efficiency of photosystem II in the light
ROI
region of interest
HL
high-light
LL
low-light
IVG
interveinal gap
SD
stomatal density
IVD
interveinal distance
SCA
stomatal complex area
SCL
stomatal complex length
SCW
stomatal complex width
CFN
cell file number
CFW
cell file width
RNAseq
RNA sequencing
qPCR
quantitative PCR
cDNA
complementary DNA
Citation
Julia, Yaapar, M. N., Supatthra Narawatthana, Christoph Lehmeier, Samart Wanchana, Thakur, V., Chater, C., Kelly, S., Rolfe, S. A., Quick, W. P., & Fleming, A. J. (2016).
Combined Chlorophyll Fluorescence and Transcriptomic Analysis Identifies the P3/P4 Transition as a Key Stage in Rice Leaf Photosynthetic Development. PLANT PHYSIOLOGY, 170(3), 1655–1674. https://doi.org/10.1104/pp.15.01624
Authors for correspondence:
Andrew J. Fleming
Email: a.fleming@sheffield.ac.uk
Attribution 4.0 International — CC BY 4.0 - Creative Commons


Comentários